Digital Biology/R
A 4-week introduction to the fundamentals of the R statistical programming language usage in the context of biological research. This is an experiential learning course where students will have private access to real virtual machines (VMs) running in the cloud. Students will collaborate with each other, human mentors, and the AI assistant Ask Stinger. After applied learning of R programming skills, learners will apply their skills to analyzing public clinical cancer data. Qualifies for the Digital Biology/R digital badge.

Course Details
Biology covers many areas including ecology, medicine, genetics, zoology, agriculture, biophysics, bioengineering, biochemistry, and many more. A recent theme in biology is the generation of a lot of biological data that needs to be analyzed by computers. For instance, DNA can now be cheaply sequenced (think 23andMe) providing insights into human ancestry and medical risk factors.
Digital Biology is a dynamic scientific discipline that utilizes computational and statistical methods for solving biological problems. A major theme in digital biology is to integrate and understand biological data generated by genome sequencing projects and other high-throughput molecular biology efforts. Digital biology tools are developed to reveal fundamental mechanisms underlying the structure and function of macromolecules, biochemical pathways, disease processes, and genome evolution.
In this course, you will receive a hands-on introduction in the R programming language using R-studio notebooks. The first half of the course is hands-on learning of R syntax including basic data structures, loops, conditionals, and I/O. The second half involves writing functional R code that parses real clinical data and performs basic statistical operations on the data.
Journey Architecture
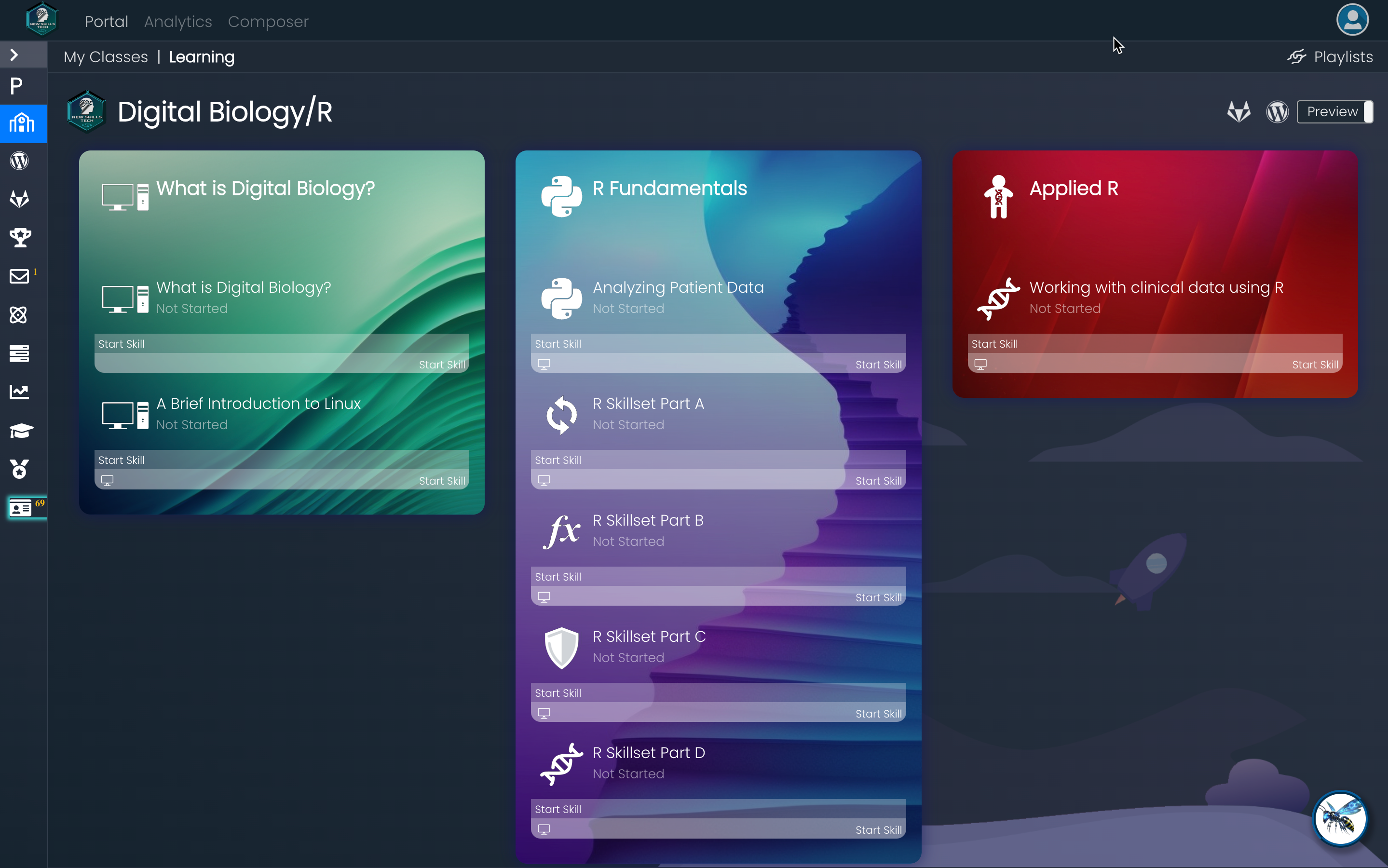
This course contains three learning paths each containing topic specific skills. Below is a screenshot of the learning paths and skills followed by a bulleted list.
The first learning path (What is Digital Biology?) is an introduction to digital biology as a specific field of study in high demand by the life science industry (public and private).
The second learning path (R Fundamentals) are hands on labs where learners interact with a real Linux VM running R-studio to understand R syntax.
The final learning path (Applied R) will test the learner’s abilities by having them write R code to explore a publicly available clinical dataset.
●
What is Digital Biology? (Learning Path)
●
What is Digital Biology? (Skill)
●
A Brief Introduction to Linux (Skill)
●
R Fundamentals (Learning Path)
●
Analyzing Patient Data (skill)
●
Data Analysis Setup (resource)
●
Analyzing Patient Data (resource)
●
R Skillset Part A (skill)
●
Creating Functions (resource)
●
Analyzing Multiple Data Sets (resource)
●
Making Choices (resource)
●
Command-Line Programs (resource)
●
R Skillset Part B (skill)
●
Best Practices for Writing R Code (resource)
●
Dynamic Reports with knitr (resource)
●
Making Packages in R (resource)
●
R Skillset Part C (skill)
●
Introduction to RStudio (resource)
●
Addressing Data (resource)
●
Reading and Writing CSV Files (resource)
●
Understanding Factors (resource)
●
R Skillset Part D (skill)
●
Data Types and Structures (resource)
●
The Call Stack (resource)
●
Loops in R (resource)
●
Applied R (Learning Path)
●
Working with clinical data using R (Skill)
Skills and Resources
Digital Credential

Earners of the Digital Biology/R BioHacker credential have successfully demonstrated experiential skills in Linux command line interface and installing genomics software. The Digital Biology/Linux badge requires 20 hours of hands-on activities and labs across 8 skills in relevant to biotech R&D.